October 27, 2020
I first discussed the concept of methylation-based epigenetic aging clocks, and how you can learn from them your true biological age, on the podcast “Is This The Most Advanced Anti-Aging Analysis & Epigenetic Diagnostic Test Of The Future?“
This is an incredibly exciting technology that has many implications in how aging is studied—with some of the most profound benefits often overlooked. If you're looking to track how fast you're aging or what your true “biological” age is, then this is about the most cutting-edge approach you can take.
Essentially, these aging clocks interpret markers of methylation on the DNA. Methylation markers are a type of epigenetic modification that your body uses to regulate gene expression. Every cell in your body has the same baseline DNA sequence; however, epigenetic expression is what gives certain cells their phenotype. That is how your heart cells become heart cells and your eye cells become eye cells.
Usually, this methylation is a way to silence gene transcription. This epigenetic regulation isn’t set in stone and often methylation markers can be added or removed based on lifestyle factors and interventions. When these markers were viewed across lifespan, it became clear that their changes might be a result of more than just a coincidence and instead reflect the actual process of aging. As a result, many researchers look at these changes to create clocks that could measure and predict aging.
So in today's article, you’ll discover everything you need to know about some of the most important impacts of methylation-based epigenetic aging clocks, why we need them in the first place, how you can incorporate them into your own approach for optimal aging, and much more.
The Need For A Great Aging Biomarker
It is no secret that chronic diseases comprise the majority of the global disease burden and are the most common cause of mortality—with 80% of adults age 65 and over having at least one chronic disease.
Despite this, there is still great heterogeneity in the health outcomes of older individuals. Some folks in their 70s appear frail and require assistance in daily routines, whereas others live independent lives and seem to escape major physiological deterioration well into old age. Why is this?
Unfortunately, predicting the factors that contribute to this wide range of outcomes has been incredibly difficult, and biomarkers that predict and track the state of biophysiological aging are much needed. During the past decade, extensive effort has been made to identify such aging biomarkers that are “biological parameters of an organism that either alone or in some multivariate composite will, in the absence of disease, better predict functional capability at some late age, than will chronological age.”
Although crude markers of biological age (telomere length, transcriptomic predictors, proteomic predictors, metabolomics-based predictors, and composite biomarker predictors) have existed for decades, no biological marker has sufficiently satisfied the requirements put forth by the American Federation for Aging Research (AFAR) listed below:
- It must predict the rate of aging. In other words, it would tell exactly where a person is in their total life span. It must be a better predictor of life span than chronological age.
- It must monitor a basic process that underlies the aging process, not the effects of disease.
- It must be able to be tested repeatedly without harming the person. For example, a blood test or an imaging technique.
- It must be something that works in humans and in laboratory animals, such as mice. This is so that it can be tested in lab animals before being validated in humans.
Moreover, the existence of such biomarkers has been questioned, because the effects of many chronic diseases are inseparable from normal aging. The rate of biological aging can also vary across different tissues, and hence it may not be feasible to assume a measurable overall rate.
The Horvath and Hannum Clocks
After years of trying to find a great surrogate method for biological age, there have been several novel clocks that meet these aforementioned criteria. Most of them center around the multi-omic molecular biomarkers such as transcriptomics, proteomics, and metabolomics.
The general consensus of the research community establishes one type of clock in particular as the most promising: epigenetic methylation clocks.
The investigation into methylation-based biomarkers really began with the following studies…
- Intra-individual Change Over Time in DNA Methylation With Familial Clustering
- Aging and Environmental Exposures Alter Tissue-Specific DNA Methylation Dependent upon CpG Island Context
- Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer
…which were some of the first to show that methylation at specific loci across the human genome changed with age. Another study showing that 88 CpG sites (where methylation of DNA occurs) from saliva were highly correlated with age stimulated the search for CpG sites that would be biomarkers of chronological age.
Building from these initial observations, investigators used supervised machine learning methods on datasets containing both DNA methylation data and chronological age information to “learn” which CpG sites in the human genome could predict chronological age with high accuracy. The predictive accuracy of the trained models in estimating chronological age was then validated in independent cohorts of subjects of various ages. Using this methodology, several epigenetic clocks have been developed.
Most notably were the clocks published in 2013 by biostatisticians Dr. Greg Hannum and Dr. Steve Horvath. Both of these clocks were able to show high age correlations (r = 0.96 for Horvath and r = 0.91 for Hannum) and small, mean deviations from calendar age (3.6 and 4.9 years, respectively).
The correlation coefficient “r” measures the strength and direction of a linear relationship between two variables. Values between 0.7 and 1.0 indicate a strong, positive linear relationship.
The Horvath clock is probably the most famous due to his subsequent work and investigations in the area. The clock, also known as “DNAm age,” is a multi-tissue predictor based on methylation levels of 353 CpG locations, whereas the Hannum clock uses only 71 CpG locations and performs best using whole blood samples. The selection of the CpG sites for both predictors was done using a similar penalized regression model, yet they only have six CpG sites in common.
Since then, several other biological age clocks have been developed. This includes Dr. Horvath’s GrimAge and Dr. Morgan Levine’s PhenoAge. All of these have different strengths and benefits as shown in the graphic below.
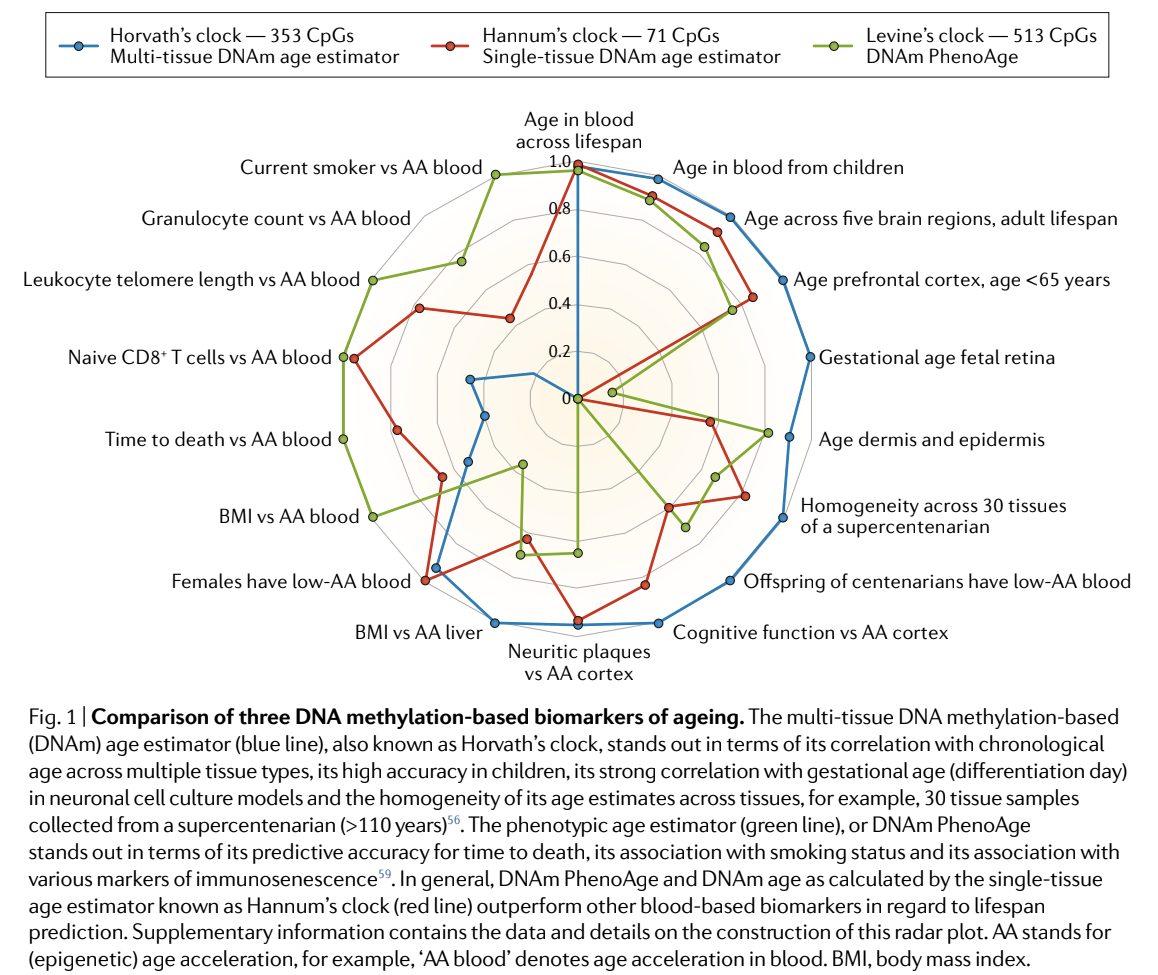
I know that graphic could be a lot to unpack, but the main takeaway is that the algorithms created from these computer learning programs all have different strengths and weaknesses due to the way they were created and the way the data was collected. Some are better at predicting time to death and others are better at predicting aging of the skin. The reasons for their strengths and weaknesses could be due to the tissues that were investigated, or the genetic locations chosen for the algorithms. Since epigenetic characteristics are different in each cell; there can be a lot of heterogeneity. While the methods to measure methylation are similar and predictable, the algorithms to interpret this data can vary greatly and the power of the algorithm depends on the outcome you want to measure.
Can These Clocks Predict Disease-Related Outcomes?
The best biological aging clock is not necessarily the one that predicts age correctly. Instead, the best biological clock should be able to predict outcomes related to the aging process.
The blood-based algorithm by Hannum and the multi-tissue algorithm by Horvath produce age estimates (DNAm age) that correlate with chronological age well above r=0.90 for full age range samples. They also exhibit statistically significant associations with many age-related diseases and conditions.
However, often the correlation to certain age-related diseases can be small to moderate. One explanation is that using chronological age as the reference, by definition, may exclude CpGs whose methylation patterns don’t display strong time-dependent changes, but instead signal the departure of biological age from chronological age. Thus, it is important to not only capture CpGs that display changes with chronological time, but also those that account for differences in risk and physiological status among individuals of the same chronological age.
Thus the investigation for better algorithms has already begun, and several have been published. This includes Dr. Levine’s PhenoAge and Dr. Horvath’s GrimAge. These algorithms have improved by becoming more like composite biomarkers including things like basic blood values and protein assay prediction.
The end result of this is an extremely accurate prediction of outcomes such as cardiovascular disease, cancer, and even death.
For instance, one study found that for each one year increase in the difference between chronological and epigenetic age (Δage), there was a 6% increased risk of developing cancer within three years and a 17% increased risk of dying of cancer in the next five years.
Additionally, some studies calculated hazard ratios for Δage (per 5 years) calculated using the epigenetic clock developed by Horvath. These were able to demonstrate a hazard ratio of 1.23 for all-cause mortality, 1.22 for cancer mortality, and 1.19 for cardiovascular mortality after adjustment for batch effects, age, sex, educational level, history of chronic diseases, hypertension, smoking status, body mass index, and leukocyte distribution.
The Next Step For This Technology
The field of epigenetics is still in its infancy and there are many upcoming developments. Several companies are investing millions into retrospective and prospective studies that are looking into better algorithms for biological age prediction, death prediction, and a myriad of disease-specific algorithms.
Beyond that, several epigenetic companies are also investigating how interventions are able to predict methylation patterns and, therefore, disease risk.
For example, TruDiagnostic (the same test I discuss in detail in this podcast) is one of these more accurate tests. To date, they have 14 approved, prospective institutional review boards looking into some of the best interventions to reduce epigenetic aging like plasma apheresis, senolytics, mitochondrial peptides, and many more. You can see my TruDiagnostic TruAge results here. (I was actually surprised to see that despite telomere testing revealing a low biological age for me, once the epigenetic results were revealed, it told a different story that influenced a few lifestyle changes I made to reduce inflammation, including less heavy heating with cooking oils, more recovery time between hard workouts, and a higher intake of life-extending compounds such as NAD and sirtuins, and other strategies I discuss in detail in Boundless).
Methylation-specific loci are also being investigated for their specific risk of disease which, unlike the DNA sequence, can be influenced by the environment, and have the potential to improve disease prediction. Recently, one epigenome-wide association study identified 5 DNA methylation loci (ABCG1, PHOSPHO1, SOCS3, SREBF1, and TXNIP) in the blood that were associated with type II diabetes. The study showed that a methylation score that combined the results from these 5 methylation loci found an association with prospective type II diabetes occurrence. It is so accurate that its predictive value within 7 years is better than HbA1C levels and fasting insulin.
While all of these developments and their implications are exciting, I believe that some of the best implications are commonly missed:
First, with these new methylation-based clocks, we now have the ability to evaluate the effects of anti-aging therapies objectively and in real-time. For instance, let’s imagine that you’re excited about a new drug and you want to know whether it really assists in extending lifespan and healthspan. Before the development of these clocks, you would have had to give the medication to thousands of patients and wait years to see if fewer of them were dying, compared to people who did not get the drug. These new clocks provide a huge shortcut. Now, you can give people medications, supplements, or lifestyle interventions and measure their epigenetic markers before and after the intervention. Even with small sample sizes, you can get a very good idea of how the intervention is working and how it is affecting disease risk in several categories.
Second, the implications of this testing get even better when you consider that it can give objective feedback in a personalized manner. If three people take the same medication, there might be varying degrees of success for each individual despite the same treatment conditions. For interventions and lifestyle changes that are difficult to quantify, we can now look at how specific interventions affect an individual directly instead of using a one size fits all approach to medical treatment.
The third implication might be even more exciting. At the moment, we are still unsure of exactly why we age. While we have narrowed this down to nine hallmarks of aging (genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication), the underlying mechanisms are still uncertain. However, it looks like these epigenetic changes might not just reflect changes due to aging, they might actually be the underlying causes of the aging process. This means that if we can figure out how to prevent and change the methylation changes that occur over time, we should be able to address the root cause of aging.
Summary
There is a saying in business management that you can’t manage what you don't measure. The same can be said for health-related biomarkers.
With the technology you just learned about, you now have the ability to measure the aging process and receive real-time and personalized feedback about how your exercise, nutrition, medications, and other interventions affect your own individualized aging processes.
As companies like TruDiagnostic (described above) start to develop further diagnostic algorithms, you will soon have the power to detect disease processes earlier. You will know which interventions are most likely to change the methylation patterns that contribute to specific disease processes such as Alzheimer's, diabetes, and, of course, which patterns will increase lifespan. There are already several trials that are looking at how interventions can affect epigenetic age. To get your own TruDiagnostic TruAge kit, you can click here and use code BEN50 to save $50.
While aging is one of the most exciting areas of epigenetic investigation, there are many other lessons that can be learned from the data gathered from looking at methylation. Together, the large-scale data collected from an epigenetic analysis can help you detect disease earlier, understand the best ways to treat disease, and learn about the underlying processes that cause these problems before any signs of health issues even show up.
If you have questions, thoughts, or comments about markers of aging and how to track biological vs. chronological age, leave them below!














Good and Useful
What if you tested positive for MTHFR mutation in both the A & C gene sets.
Just looking to slow down the aging process